The Regulation of Alternative Pre-mRNA Splicing
Twenty years ago, the sequence of the human genome, with its relatively low gene number, highlighted the question of how organisms produce the many functionally distinct proteins needed to sustain their complex development. Alternative pre-mRNA splicing is a major contributor to protein diversity, allowing a single gene to express multiple spliced isoforms that serve different functions. Despite now being able to quantify the breadth of splicing regulation and knowing some of the many roles splicing plays in the control of gene expression and cellular function, our understanding of the molecular mechanisms that alter pre-mRNA splicing patterns is still limited.
Our group works on a range of projects related to the control of pre-mRNA splicing in neurons and other cells. We aim to determine the mechanisms of action of splicing regulators, as well as to understand their roles in development and cell biology. We have long-standing projects focused on two families of regulatory factors: the polypyrimidine tract–binding proteins (PTBP1 and PTBP2), and the Rbfox proteins (1, 2 and 3). Each of these RNA binding proteins controls the splicing and expression of important gene transcripts in neurons and in other cells, and each provides a biochemical paradigm for examining mechanisms of splicing regulation.
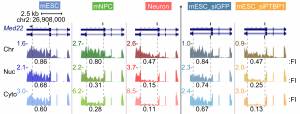
In support of our mechanistic and biological interests, we develop and apply genomewide methods to assess the programs of gene regulation mediated by RNA binding proteins. Variations of RNAseq analysis are used to define altered splicing across the transcriptome, while iCLIP-seq and other binding assays identify target transcripts and binding sites of splicing regulators. These studies provide large numbers of interesting protein/RNA interactions for further study. In collaboration with our computational biology colleagues, we use these datasets to examine global regulatory mechanisms, including defining the RNA binding specificity of proteins and measuring intron excision rates.

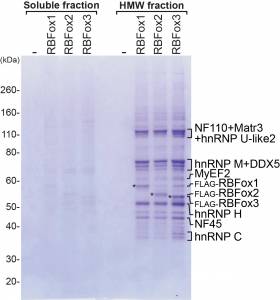 In mechanistic studies, we use biochemical methods, tissue culture, and in vitro splicing systems to examine the molecular interactions of splicing regulators and analyze how they can alter spliceosome assembly. We isolate RNP complexes assembled onto regulated exons and characterize their architecture, using chromatographic, molecular crosslinking, and mass spectrometric methods. Following this approach, we have defined the interactions of the regulatory protein PTBP1 with the spliceosome that mediate its splicing repression activity. We also showed that the Rbfox proteins engage with a novel higher order protein complex of multiple splicing regulators, whose activities are modulated by their interactions with Rbfox. Collaborating with structural biology groups, we work to define the structure of these proteins and their RNA binding domains.
In mechanistic studies, we use biochemical methods, tissue culture, and in vitro splicing systems to examine the molecular interactions of splicing regulators and analyze how they can alter spliceosome assembly. We isolate RNP complexes assembled onto regulated exons and characterize their architecture, using chromatographic, molecular crosslinking, and mass spectrometric methods. Following this approach, we have defined the interactions of the regulatory protein PTBP1 with the spliceosome that mediate its splicing repression activity. We also showed that the Rbfox proteins engage with a novel higher order protein complex of multiple splicing regulators, whose activities are modulated by their interactions with Rbfox. Collaborating with structural biology groups, we work to define the structure of these proteins and their RNA binding domains.
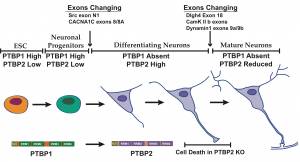
In the second arm of our studies, we seek to understand the role of splicing regulation in areas of cellular and organismal biology, including cancer, innate immunity, neuronal development, and neuronal physiology. To assess the function of splicing regulators and the specific mRNA isoforms they create, we apply genome editing, gene transduction, and pharmacological approaches in mouse, tissue culture and stem cell models. Collaborating with groups who specialize in neuroscience, immunology, and cancer biology, we identify roles for splicing regulation in development, physiology and disease. In past work, we showed that the PTBP1/PTBP2 proteins control expression of genes essential to neuronal maturation. We demonstrated that Rbfox proteins control neuronal excitation in the hippocampus through the posttranscriptional regulation of synaptic proteins. We found that delayed splicing of particular introns in inflammatory genes determines the kinetics of their expression during the innate immune response of macrophages.
Over time and with diverse collaborators, we have been excited to explore many other aspects of RNA molecular biology and the function of RNA binding proteins. Questions of special interest have included:
- How RNA binding proteins affect miRNA biogenesis and translation.
- How neuronal excitation and calcium signaling alter splicing patterns.
- How the nuclear/cytoplasmic compartmentalization of RNA affects gene regulation.
- The role of protein/RNA condensates in splicing regulation and gene silencing.
- Development of predictive computational models of RNA binding and splicing regulation.
- Identification of small molecule modulators of splicing.
- The role of the Myc oncogene in driving cancer specific splicing programs.

