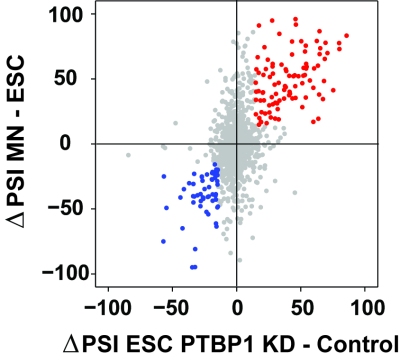
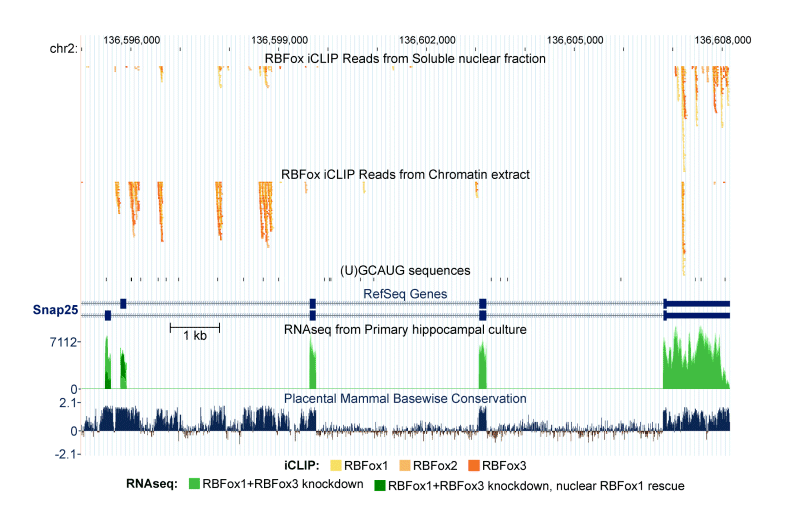
In support of both our mechanistic and biological interests, we make extensive use of genomewide methods to assess the broad programs of gene regulation mediated by RNA binding proteins. RNAseq analysis is used to define altered splicing across the transcriptome. iCLIPseq is used to identify target transcripts and binding sites of splicing regulators. These studies provide large numbers of interesting protein/RNA interactions for further study. In collaboration with computational biology colleagues at UCLA, we also use these datasets to examine general questions of regulation, such as defining the RNA binding specificity of proteins and measuring in vivo intron excision rates.